This year, CooperSurgical brought a message of quality to the ESHRE 2020 virtual congress. On that topic, Drs Josh Blazek and Tony Gordon spoke about the increasing quality of PGT over time. They reviewed the technological advances that have made PGT improvements possible and demonstrated how these gains in quality impact patient care.
Dr Blazek focused on the molecular and bioinformatic methodologies behind PGTaiSM 2.0 Plus. He began by reviewing how far PGT has come over the past three decades—from FISH, to aCGH, and recently to next-generation sequencing (NGS). He also gave a quick primer on single nucleotide polymorphisms, or SNPs: those common, single-letter changes that occur approximately every 400-500 base pairs in the human genome.
SNPs are common, presenting in at least 1% of the population, and are typically benign. At any given locus, an individual can have the more common SNP (called the major isoform or allele A) or the less common SNP (called the minor isoform or allele B).
No two people (except identical twins) have the same pattern of SNPs, so an individual’s unique SNP pattern can be used as an identifier in genomic analyses. SNP assessment from NGS has historically been achieved by one of two methods: the more extensive (and expensive) whole-genome sequencing method, and the more economical targeted sequencing method (Figure 1).
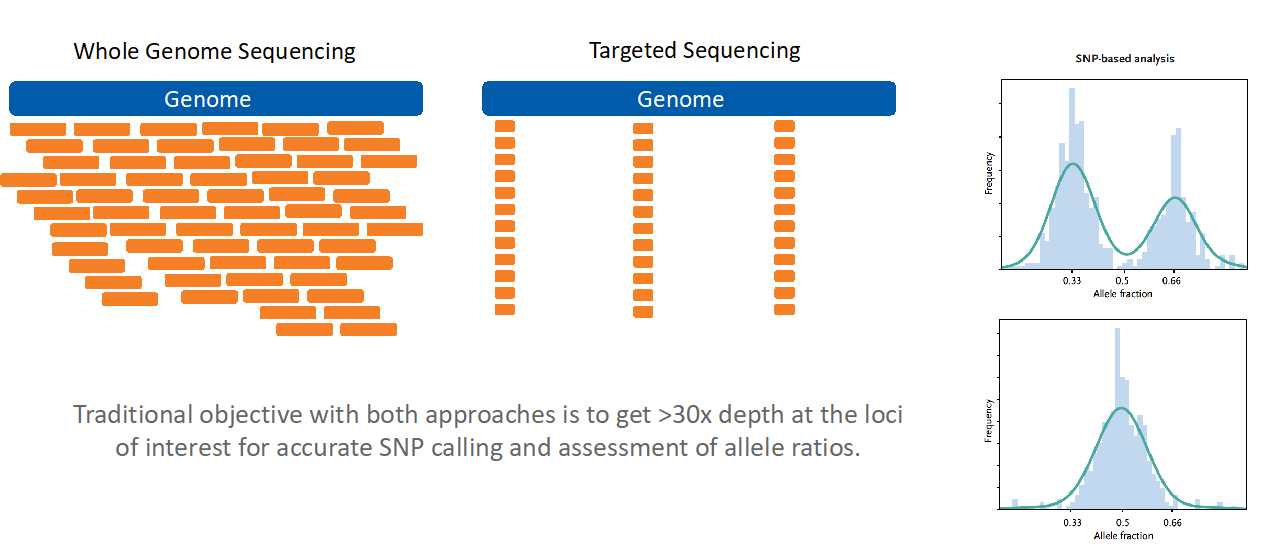
Figure 1
Dr Blazek outlined the R&D team’s thought process behind their most recent NGS enhancements. They had originally turned to artificial intelligence to streamline the reporting process while eliminating subjectivity and transcription errors.
They had focused on optimizing copy number variant (CNV) calling as much as possible. At that time, they were not attempting to create a test that could assess SNPs and CNVs simultaneously. Therefore, further innovations needed to be made once they decided to add SNP calling to the platform.
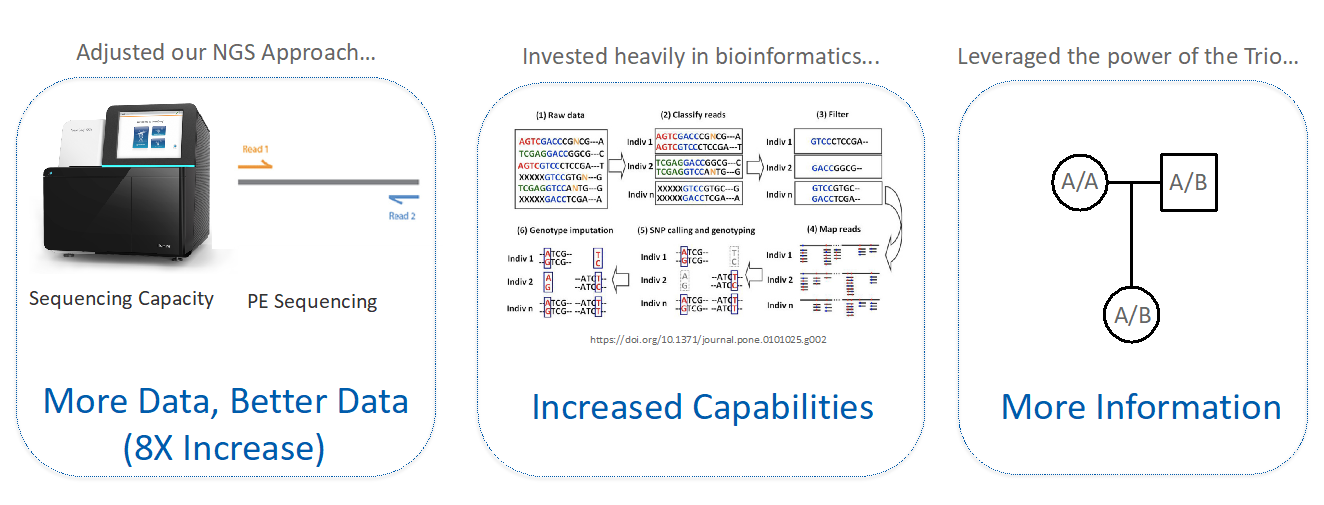
Figure 2
To achieve simultaneous CNV and SNP calling, the team adjusted their NGS approach to generate higher-throughput and higher-quality data; invested in bioinformatic upgrades; and leveraged the family trio. They increased data output eightfold by adopting paired-end sequencing and upgrading to the Illumina NextSeq sequencing platform.
They entirely reworked the bioinformatics pipeline to utilize SNP data as well as CNVs. They then were able to use that SNP data to assess mother-father-embryo trios and add entirely new functionality to the PGTai test (Figure 2).
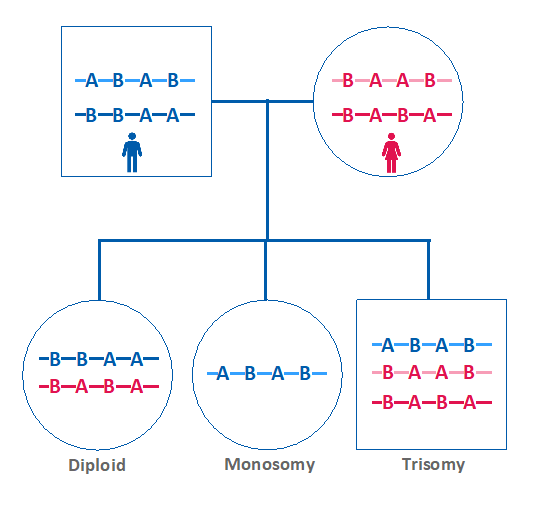
Figure 3
In addition to the traditional NGS CNV calling, the new improvements enabled simultaneous SNP analysis as a secondary measure of embryo ploidy. This is achieved by comparing the maternal versus paternal dosage of SNPs.
If the maternal and paternal SNPs are present in equivalent amounts in the embryo sample, then the embryo is expected to be euploid. However, if one parent’s SNP pattern is over or under-represented in the embryo, that indicates the presence of aneuploidy (Figure 3).
SNP analysis is crucial for a key update for PGTai 2.0: the ability to detect haploidy and all forms of triploidy. NGS alone cannot identify aneuploid embryos with 23,X or 69,XXX karyotypes. SNP analysis is required to distinguish these findings from 46,XX embryos. For example, a 69,XXX triploid embryo would be identifiable using SNP data because one parent’s SNPs would be overrepresented for every chromosome (Figure 4). As 1-3% of all IVF embryos are expected to be triploid, this represents a substantial step forward.1
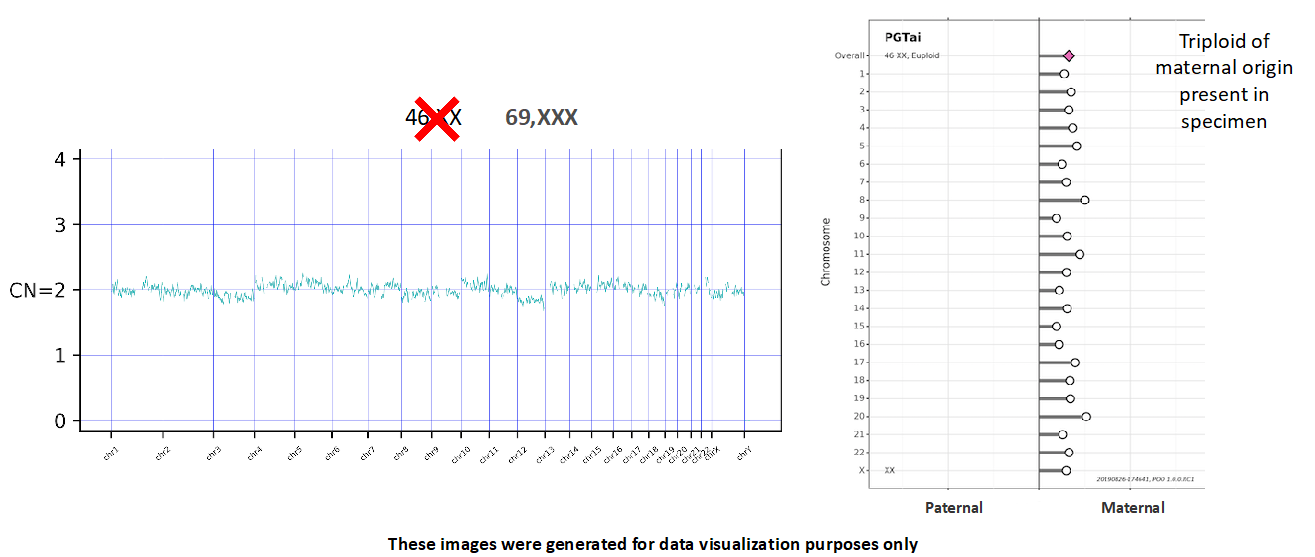
Figure 4
Furthermore, if parental samples are provided for analysis, SNP calling can determine whether an embryo’s aneuploid finding originated in the egg or the sperm (Figure 4). This can help patients and providers determine whether to use donor gametes in a future cycle.
It’s Not Over Yet: Upcoming Features
Dr Blazek concluded by announcing that his team has been at it again. They are currently finishing another update that will utilize PGTai 2.0’s SNP data to identify a genetic match between the embryo sample and the submitted parental samples, i.e. a parental quality check (parental QC).
To design this newest analysis, Dr Blazek and his team utilized a training set of 132 trios from 27 PGT-M families, including 14 consanguineous families. The trios were analyzed on the PGTai 2.0 SNP platform in numerous permutations with incorrect maternal DNA, paternal DNA, or both.
They then validated this algorithm with an independent test set of trios. The independent validation identified a 99.7% accuracy rate with 100% sensitivity and 99.4% specificity (Figure 5). Dr Blazek anticipated that this technology will impart peace of mind for clinicians and patients.
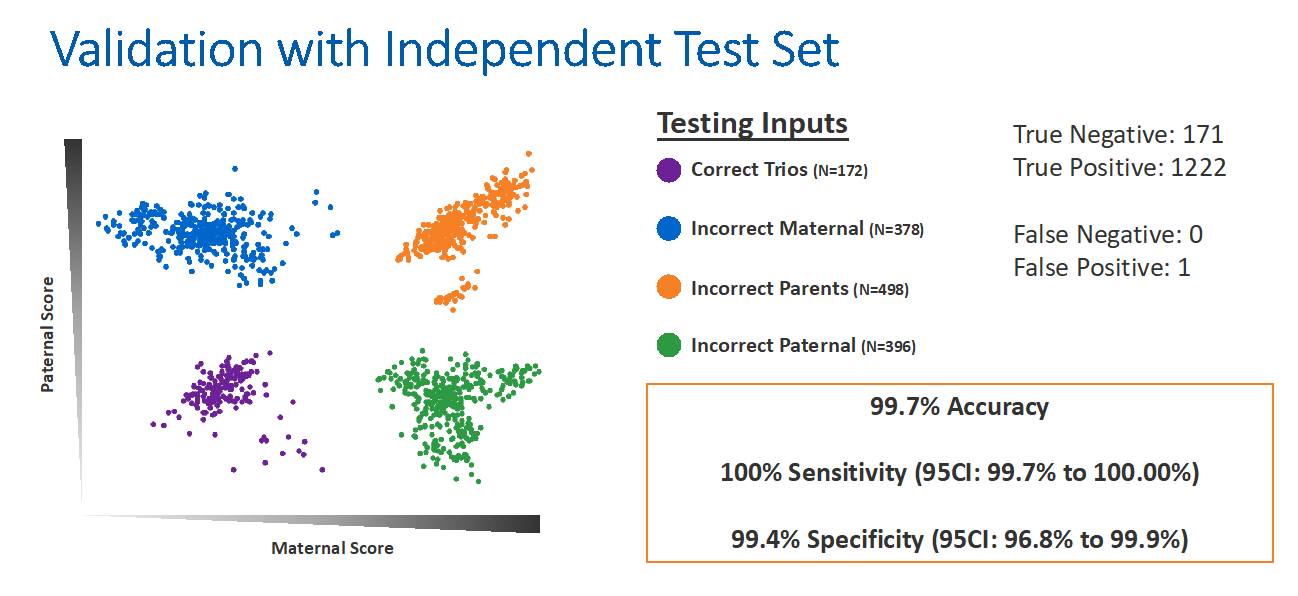
Figure 5
CooperSurgical’s Chain of Quality
Dr Gordon rounded out the session by speaking briefly about CooperSurgical’s ‘chain of quality’, which included RI WitnessTM, PGTai, and PGTai 2.0. He explained that RI Witness is a robust laboratory electronic witnessing system that prevents mismatch errors, streamlines workflows, and comprehensively documents laboratory actions.
Each year, an increasing number of PGT cycles are performed all over the world, which places a greater and more complex burden on the laboratory workload. Therefore, the accommodation of these additional steps in laboratory security protocols is imperative. Using RI Witness while biopsying, vitrifying, or prepping for transfer helps safeguard against sample misidentification.
PGTai is one of the most accurate platforms for identifying euploid embryos without human subjectivity and transcription errors. It is also the most sensitive platform for mosaic calling. PGTai 2.0 identifies haploidy and triploidy and provides 2PN verification. It can also identify the parental origin of aneuploidy and, soon, parental QC. Some of these features require parental cheek brush swabs and can be assessed both prospectively and retrospectively for all samples run on the PGTai 2.0 platform.
Quality is a Team Effort
The speakers emphasized that quality is a team effort. In IVF, quality is dependent upon cooperation and collaboration between the clinician, the IVF lab, and the reference lab. Physicians, nurses, embryologists, and geneticists must rely on each other to ensure quality treatment protocols, gametes, embryos, biopsies, and genetic testing results. Every person in this process has a role to play as we all strive to achieve the ultimate objective: to help patients achieve their goal of a successful pregnancy and a healthy child.
Reference
1Nietzel, D, Walters-Sen, L, Alouf, C, Robinson, K, Zhu, M, Faulkner, N. Clinical experience utilizing single-nucleotide polymorphism data captured by FAST-SeqS to reduce the transfer of polyploid embryos. Fertil Steril 2019, 111(4):e17.
Jenna Miller, MS, CGC, is a board-certified and licensed genetic counselor. She holds a Bachelor of Science degree in Genetics and Biotechnology from Brigham Young University and a Master of Science degree in Human Genetics from Sarah Lawrence College. Jenna began her career at the biotech startup Recombine by providing genetic counseling services to patients and physicians. She then moved into clinical diagnostics, and eventually lead CooperGenomics’ carrier screening clinical diagnostics team. Jenna has been CooperSurgical’s clinical science liaison since 2017. In this role, she travels North America educating healthcare providers about genomics within the ART field. Jenna is passionate about genomics education, informed consent, and ethical approaches to genomic testing.
Watch the Recorded Symposium
Genomics Products and Services
To find out more about our genomics products and services click here.
ART Imitating Life: the chicken or the egg?
Read Jenna Miller’s review of our second ESHRE 2020 Symposium here.

 My Clinic is in the United States
My Clinic is in the United States My Clinic is in Canada
My Clinic is in Canada

