Find out about the great work we have been undertaking in PGT-A, the features and benefits of this type of testing, our PGTai technology platform results and our upgrade PGTai 2.0 which is available in US labs from September 2019.
Webinar Transcript: Explore PGT-A using our revolutionary PGTai technology platform
In this webinar, CooperSurgical’s VP of Global Genomics Business Development, Tony Gordon PhD discusses the work Cooper Genomics has been undertaking in Preimplantation Genetic Testing for Aneuploidies (PGT-A), the features and benefits of this type of testing, our PGTai technology platform results and our upgrade, PGTai 2.0.
Today, I’m going to tell you a little bit about some of the work we’ve been doing here at Cooper Genomics to look at PGT-A (formerly PGS). This is the technology that we’ve been developing here to try to improve the quality of this technology and try to bring some new features and benefits to our IVF customers and, hopefully, our patients.
The talk will be split into three parts:
- The first part is just a little recap as to why we’re here, doing PGT-A; fairly simple, fairly straightforward.
- The second part is to tell you about our technology that we’re using right now for our customers – that’s PGTai (AI standing for Artificial Intelligence). I’ll go through that and look at the results we’ve had since we launched that technology ten months ago.
- Then I’ll go on to tell you a little bit about the technology that we have developed and we’re releasing next week (on September 3rd). All samples that are sent to Cooper Genomics from September 3rd will be on our new technology, which is PGTai 2.0.
Cooper Genomics has been around in this area for as long as pre-implantation genetic testing has existed. The very first PGT-M case (as we used to know it, ‘PGD’) was done by one of our founders. The very first PGT-A (or ‘PGS’ as we used to call it) was done by one of our other founders. And we’ve done more than a half a million procedures in PGT over the years so we, clearly, have the most experience in this area. What we’re now trying to do is push the technology and use our experience and our knowledge to see where we can go.
There are three areas of PGT. I’m going to talk almost exclusively about PGT-A (aneuploidy testing) or, as it was known before, PGS, CCS and a bunch of other acronyms. That’s screening for chromosomal abnormalities and embryo biopsies. It’s an option to all IVF patients and there are some really good data that have just been coming out from SART (Society for Assisted Reproductive Technology) and some other areas that shows it improves IVF success rates, even per intent to treat. I’ll not talk about that data but that’s some really cool data that might be in a subsequent webinar.
PGT-SR is similar but different. That’s where you know that the couple coming in are carriers – one of them, usually – of a chromosomal rearrangement, so you’re looking for changes in the chromosomes associated with that chromosomal rearrangement because if you get an unbalanced embryo then it’s very, very likely to end up in a miscarriage. So the technology I’m going to talk to you about today – PGTai and PGTai 2.0 – is essentially what we’re going to use for PGT-SR (except, with SR, we just need some additional information for the chromosomal rearrangement that the couple have).
What I’m going to talk very little about is PGT-M (or, as we used to call it, ‘PGD’) except to say that this is a technology that we’ve been using for many, many years and we use a technology called “SNP Arrays” for this – I’ll come back to that later on in the presentation.
What we do is not in isolation. What we do is associated, and in collaboration with, IVF clinics, embryologists and doctors. So, in order to make the PGT-A process a success, you need:
- Good quality IVF
- Good quality embryo biopsy (without good quality embryo biopsy, what we do is not-quite meaningless, but is very difficult to do)
- And you need vitrification.
Only once you get to this part of the process does the sample come to the lab for us. We try to get results wherever possible and then give you the results and, hopefully, you can then make a decision on which embryos to transfer.
Cooper Fertility and Genomics Solutions (although I’m talking just about the Genomics today), obviously can help through the IVF process, embryo biopsy (we have great embryo biopsy workshops in our Center of Excellence, Livingston New Jersey) and the whole process there. But let’s stick to PGT-A.
PGT-A
The premise of PGT-A is fairly simple. If you have an embryo biopsy with the correct number of chromosomes (that’s 46), that is called euploid. If you implant an embryo with that correct number of chromosomes, it’s likely to have a higher success rate than one with the wrong number of chromosomes. Even implanting euploid embryos doesn’t guarantee success but randomized control trials and the analysis of SART data shows that it does improve outcomes.
However, if you have the wrong number of chromosomes [you have a lower chance of success.]
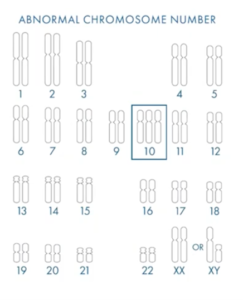
This is a fairly simple example. You can see we have 47 chromosomes in this embryo biopsy, so this is a meiotic error (this occurs in the egg or the sperm). All the cells have one extra chromosome (in this instance, of chromosome 10) and I can tell you, as a cytogeneticist, that there has never been a healthy live birth with a 47 triple 10 genetic abnormality. So you know that, if you put that embryo back, it’s not going to succeed. So we’re screening for the correct number of embryos.
Aneuploidy – as we know – increases with maternal age and it’s one of the main causes of infertility, so we’re trying to avoid those aneuploid embryos, particularly with advanced maternal age. If you implant those aneuploid embryos, they will quite often result in failed implantation. There’s a huge amount of data now that shows that you have increased miscarriage rates with aneuploid embryos. And, finally, it can cause genetic disease; so, if you implant an embryo with trisomy 13, or trisomy 18, or trisomy 21, you’ll get a genetic disease like Down Syndrome, Edwards Syndrome or Patau Syndrome. So, that’s the premise.
Up until a few years ago, it was fairly simple because we thought gaining an extra chromosome or losing a chromosome was bad. However, technology’s moved on and our understanding of embryo biology has moved on, and now what we’re seeing is embryo mosaicism. But, what happens in mosaicism – put simply – is you have two or more cell lineages in an embryo.
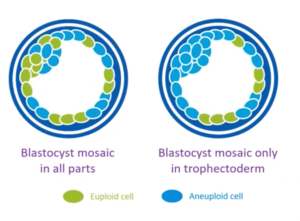
You can see here (on the left), you have an embryo which has euploid cells in green, and aneuploid cells and blue. On the right, a slightly different one with aneuploid cells in blue, euploid cells in green. And, if you take that biopsy, you now have to not just count whether there’s a gain or loss of chromosomes, you have to count whether there’s partial gains or losses. And that’s more challenging for us doing the genetic testing, and we’ve had to improve our technology to be able to do that.
Mosaic embryos, though, are really interesting because there’s been a small but growing amount of evidence that showed that they implant less, they miscarry more, but they can sometimes lead to live births. So sometimes, if you have no euploid embryos, you might not want to discard a mosaic embryo, but consider whether it’s something suitable to transfer because many centers are now seeing live births after mosaic embryo transfer.
What we’re trying to do is improve the information we can give about levels of mosaicism as well. What I would say is that mosaicism does occur naturally and some of us out there probably are mosaic. So, the PGT-A results that we generate are in four classes. It used to be nice and simple (euploid vs aneuploid). But now, if the lab wants to know, we now will tell them whether they’re mosaic.
In euploids, all the chromosomes are normal; high chance of success rates. Aneuploid, all the cells are abnormal and it has almost no chance of a healthy live birth. In between, you can get mosaics – low-level and high-level. Low-level, we class here at Cooper Genomics as 20 to 40% abnormal cells; and high-level, 40 to 80% abnormal cells.
What we see is, with the lower-level mosaic biopsies, they have not such good outcome as a euploid, but better than high-level. The high-level have a better outcome than aneuploid, but worse than the low-level mosaics.
So, we’re trying to give the clinic the best chance of finding a euploid or a mosaic to give the patient the maximum chance of getting something to transfer.
This is where we were up until less than a year ago. We were using technology that was originally developed – and I was one of the people that developed this back in the day – back in 2014. Here we are in 2019! And, just to kind of give you some kind of comparison, in 2014 we were using iPhone 5s; now we’re on iPhone 10s and iPhone X! The technology has moved on; five years is a long time in the high-tech arena and we were fixed on one test. What we’ve tried to do is develop our own custom test that enables us to do more things in a different way.
PGTai
This is the test that we’ve been running since November 1st last year. AI stands for Artificial Intelligence, and it’s been a ground-breaking platform in pre-implantation genetic testing. It’s a first-of-its-kind mathematical algorithm and it allows us to harness big data, artificial intelligence and machine learning to improve our results, to reduce noise, increase signal and maximize sensitivity and specificity.
When we say Big Data, we’re doing tens – if not hundreds – of thousands of embryo biopsies and we’re getting hundreds of thousands of next-generation sequencing reads, so we’re talking about data in the billions of data points and that’s where you can start to use the power of this kind of technology.
This enabled us to go back and think about how we were actually doing PGT-A and how we’re using the technology and try to go back with a clean sheet of paper to see how we could improve the technology.
What we did was different from what the softwares that have been used in the past were doing. In this technology you have to use something as a baseline for ‘normal’ and then you can call ‘abnormal’ from that. Previously, software – including the software that I’d developed in the past with various teams – would use cell lines. Cell lines, obviously, aren’t embryo biopsies and they were quite small cell line cohorts. We took embryo biopsy data and, for our ‘normal’ – for our baseline – we said we want embryo biopsies that resulted in live births in sustained pregnancy outcomes. That became our baseline for the AI algorithm and then we validated using sequencing data from 10,000 embryos. The poor guys in R&D looked at that manually and looked at it via AI, and the AI did about 7% better in terms of accuracy of calling.
So, what this is allowed us to do is – for the first time – go away from manual, subjective interpretation and just use statistical calling for PGT-A. That is robust because you get the same calling every time, it’s accurate because you’ve got the same calling every time, and it removes subjectivity and avoids other errors in the process.
In the past, we’d receive a biopsy from an IVF clinic, we’d amplify that DNA about a million times, then we put that DNA onto a next-generation DNA sequencer and we would end up with a number of sequence reads. We would take that and, strangely, convert it into an image. [The line in that] image goes up and down for gains and losses and some of our scientists would then have to go through each of these images and call a gain or a loss. We’d have to have two initial independent scientists and then a third independent scientist to look at the two independent scientists’ [work], and then we’d have to put it into a report. You can see that it is a pretty high human endeavor and (we do our absolute best but) there is a risk of getting that wrong. Also, it’s a bit like embryo biopsy grading; each person might have a very slightly different view of an embryo biopsy grade. I used to think of it as [revealing] the pessimists and optimists; the optimists would say, ‘yeah, I think that’s normal. I really want to give them the best chance of having a euploid embryo for transfer’ and another person would say, ‘I can see a tiny blip there. Maybe that’s something that we need to call’. Now, we don’t look at images; we just use the stats to give us the results.
We do exactly the same processes before we do the DNA amplification on the DNA sequencer but now it’s the AI algorithm that counts the chromosomes, looking at the thousands of human biopsy samples for the normalization that produces the reports. It enables us to use Big Data and avoid subjectivity.
PGTai Platform Observations
In this section I’m going to share some of our observations we saw in the first six months of using this, compared to the previous ten months using the subjective, old technology.
The thing that’s been really interesting is that, with this system, we’ve been able to reduce the noise, improve the signal and make things much more consistent. That’s resulted in more euploid embryos being reported and an increasing number of patients with at least one euploid per cycle Just to put that into a number context, in the first six months that was 1,300 more euploid embryos being reported, which is really good because that gives our clinicians and IVF labs the chance of having more embryos to transfer for patients, which is our aim.
Looking at the numbers tells you a little bit how about how this has worked. We see a 7.7% relative increase in euploid embryos. That’s not 7.7% of all embryos but, say you have a 50% embryo euploid rate, 7.7% would be a 3.85% increase in total numbers (or 7.7% relative). So, we’re seeing maybe three or 4% of embryos that are now called euploid than would have been [previously], out of the total.
Where did they come from? Because we have a much better idea of the noise, we see a reduction in mosaic embryo reporting. 21% sounds like a big number but we only ever had about 10% of embryos that were mosaic so it’s 21% of 10% – so about 2% of embryos. Again, we have a reduction in aneuploid [of 4.2%]; these are the ones there were aneuploid and now generally called ‘high-level mosaics’. So, in all three categories we’re seeing changes.
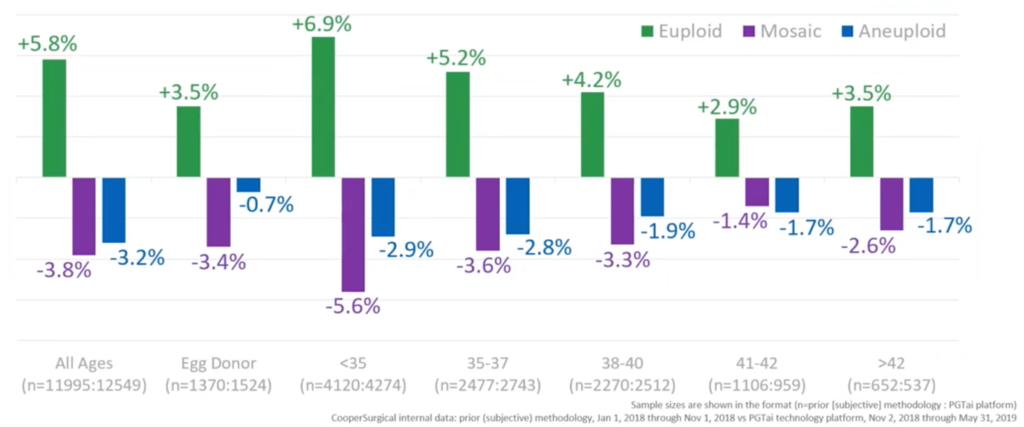
If we break that down by different groups and ages, we can see how that works. In green, we have the changes in the numbers of euploid embryos. In purple, the changes in relative numbers of mosaic embryos. And, in blue, the relative number of aneuploid embryos. And, overall, we see that change – a gain of euploids, loss of mosaics and loss of aneuploids. Interestingly, you see that gain in the number of embryos called euploid generally decreasing with age and that’s just because there’s less and less embryos that can be called euploid with age because most of them are clearly aneuploid. We’re not really sure why there’s a difference between donors and the under 35s but it’s something we’re looking at right now.
The other thing which is pretty cool is seeing how many cycles now have a euploid transfer where there wouldn’t have been. About 4% of overall cycles now have one euploid available for transfer – so it’s about one in twenty-five cycles – but still a pretty good number and very much statistically significant.
So this is PGTai, and this is what we have been running for the past ten months.
PGTai 2.0 Platform
But, when we started doing PGTai, we knew it was just the start of what we were doing because, as soon as we moved from our old technology, we were able to move on and not just change the way we do our analysis, but change the whole way we do our next generation sequencing.
Previously, we’d use small sequencers; now we’ve moved to the big generation sequencers that gives us a lot more options in the way we can do PGT and, hopefully, improve results for our clinics and, subsequently, our patients.
PGTai 2.0 still uses PGTai – the algorithms built from the biopsy data from 1,000 live births [and therefore] highly validated. But now, we have 10 times more sequencing. So, to give you some kind of indication, that’s ten times more than anybody else and up to fifty- to a hundred- times more sequencing than some other labs do. So that’s a pretty huge amount of sequencing.
Not only have we improved the amount of sequencing we do, we’ve also improved the way that we sequence. Now, instead of reading the sequence once, we read the sequence from both ends of a DNA molecule, and that tells you exactly how big the molecule is and where it is in the genome i.e. whether it was unique from the embryo. One of the things you sometimes get is duplication in the whole genome amplification or duplication in the sequencing. So this sequencing technology really cleans up the noise of the PGT-A. That’s really cool.
The other thing that we are doing, is adding SNPs to our tests. If you remember back to the very first part of this presentation, I said that we were using PGT-M for a lot of tests and we use SNPs. The way that we use SNPs back in PGT-M is using technological SNP arrays, which is quite an old technology but really good. Its really good for following the inheritance of chromosomes through generations to avoid affected chromosomes. And, when we’ve been doing PGT-M in the past, we split samples and we do PGT-A and SNP arrays. But what we wanted to do with this technology was to combine these technologies together; SNPs and copy number counting using the DNA sequencers to give the best possible tests. That’s a real tough challenge because, generally, to do SNPs using sequencing, you need to do really focused amplification to get the coverage. Doing global coverage of SNPs, at the amount we sequence, there’s a lot of papers out there that discuss this and all those papers tell you that you can’t do it. But we have found a way to do it. Now we have a test that has two completely independent methods for aneuploidy detection and calling; a primary method of next-generation sequencing and a secondary method of SNPs. This is PGTai 2.0.
The conversations about artifacts and ‘is what you see an artifact of the PGT-A processing’ is really killed off with these independent methods. It also – as an aside – lets us analyze all forms of ploidy including: haploidy and triploidy (previously we’ve been able to do male triploidy using NGS but also now female triploidy). And [we can also analyze] the origin of the aneuploidies – whether they were maternal or paternal.
To give you a little bit of a background; a ‘SNP’ (Single Nucleotide Polymorphism) is a single base pair change. They occur all the way through our genomes – we’re looking at probably between four- and five- million in each one of us – and they result from natural variation and evolution and reproduction. The majority of SNPs have no effect but they are the things that make us slightly different.
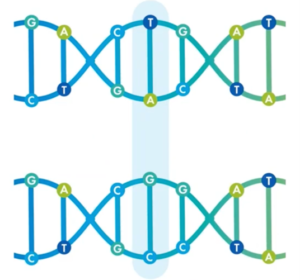
You can see here, the top strand is a TA; the bottom strand (highlighted in blue) is a GC. This is a single base pair change; we call them SNPs or, sometimes, Small Nucleotide Variation – SNV. This is what we’re able to now measure with PGTai 2.0.
We have unique patterns from Mom and Dad that are mixed together in the embryo biopsy and you have what is known as A and B, which are the major and minor alleles; the higher and lesser frequency of each SNP. Without going into too many details, we’re able to use the global assessment of SNPs to look for things like where we only see one SNP – monosomy – or three SNPs – trisomy.
Although we don’t actually look at the graphs anymore for PGT-A (it’s all statistical), it’s kind of nice just to show you some graphs just so you can get a little visual interpretation here.
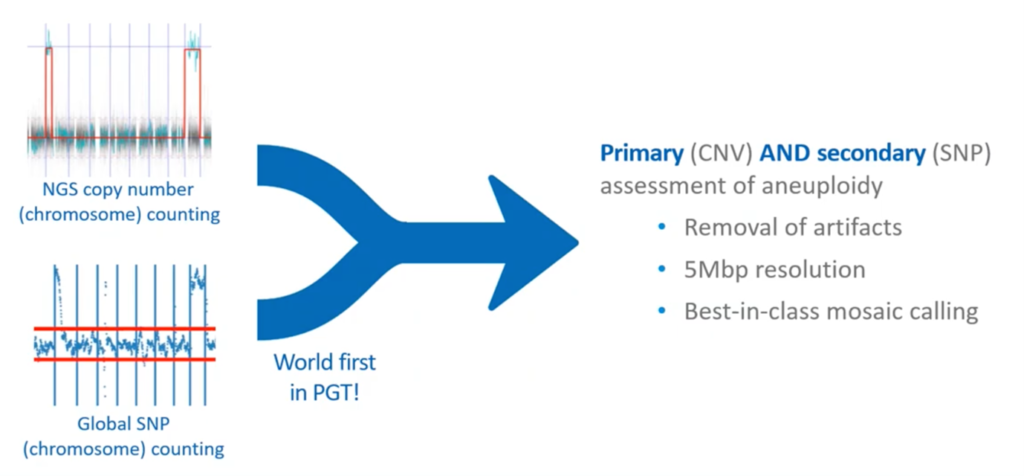
On the left here at the top, you see a copy number counting using sequencing. This is just putting the reads from the sequencer in to see how many samples we have. On the left, you can see there’s a partial chromosome gain and, on the right, a full chromosome gain.
Below, [you can see] the same DNA, but run through our global SNP counting. You can see, on the left, it also shows the segmental change (and that’s different because this SNP technology has the same resolution as the NGS) and a chromosome change. So, combining these two (primary analysis by CNV, secondary with SNPs), we have a joint aneuploidy assessment. We can remove artifacts, we have a 5Mbp resolution, and a best-in-class mosaic calling.
PGTai 2.0 Plus
In addition to the test that we have, we also have an optional, additional test. You may remember I told you that SNPs were really good at following the inheritance of chromosomes, whether that’s for PGT-M or to look for the origin of aneuploidies. This is an optional extra test and we call it ‘PGTai 2.0 Plus’.
You need parental swabs from Mom and Dad and those swabs can be sent to the clinic or the patient. It is an additional cost but it can be done as a reflex test. It doesn’t have to be done when the 2.0 test is done, it can be done retrospectively, and it can be done at any point after the 2.0 test.
The reason why we’ve done this is because there’s some interesting things you can do with looking at the inheritance of aneuploidy. This is to see where the aneuploidy has come from; meiotic with whole chromosomes, or meiotic with those segmentals.
It’s important that we can do smaller stuff as well as the whole chromosome stuff. The whole chromosome stuff is primarily maternal; over 90 percent of the time. You can generally tell, just by looking at maternal age, how much aneuploidy is going to be maternally derived, but even then, there’s still some value because sometimes it could be paternal – an indication of male factor.
The one that’s more interesting is really the segmental aneuploidies because, if you see quite a lot of these, we should really look at them. For the first time, we’ve seen a genetic chromosomal change in embryos which is majority paternal and these segmental aneuploidies could be a real indication of male factor.
Additionally, parent of origin is useful for the couples just because you might have a cycle with no embryos to transfer. It also gives patients closure, or it helps decisions for things like gamete donation, so we’ve added that as an additional test.
In conclusion:
- PGA 2.0 is a combination of using sequencers to copy count chromosomes (copy number variation) but also using sequencers to look for SNPs, and adding these two together as two independent assessments of ploidy.
- We were able to do that because we’re now generating ten times more data than we did before.
- It detects all forms of triploidy, as well as haploid and some forms of other ploidy
It allows for parent of origin testing and it really bolsters the embryologist’s 2PN validation because it will see all forms of ploidy.
Support and Quality Assurance
We’re not just here for the pure testing. One of the things that we’ve been doing with PGTai is clinic report cards. If you’re a CooperSurgical customer we will look at your euploid rate, we will give you results – if you want – per clinician (what their euploid / aneuploid rates are), we’ll look at mosaic levels and we can also compare those results from a single clinic with the global Cooper Genomics results.
We also offer genetic counselling. That’s not something we routinely offer for PGT-A but, obviously, the test is becoming a little bit more complicated and we’d be happy, in many circumstances, to assist much more if you want to know about PGTai 2.0 through the all-patient results.
We also have a very cool technology called EngagedMD and this is a tool that patients can use online before they come to the clinic to give them some sort of introduction to the area and help with many of the difficult questions that they might come up with.
Finally, although we’ve authored many, many scientific publications and abstracts we’re always open to partnership and collaboration and we’d love to be in touch with you if you want to find out more about PGTai 2.0.
About the Presenter: Tony Gordon PhD
Dr Gordon is a PhD molecular cytogeneticist with over 20 years’ experience in molecular diagnostics.
He is currently the Direct of Clinical Operations for CooperGenomics UK labs, and is leading the CooperGenomics global business development – outside the US. Dr Gordon is also a UK State Registered Clinical Scientist and is a Fellow of the Royal Society of Biology.
Read more about Tony Gordon PhD.
Useful links
If you would like to explore any of the products / references mentioned in this article, please click on the links listed below:
- Preimplantation Genetic Testing for Aneuploidies (PGT-A)
- PGTai Technology Platform
- PGT-M
- SART (Society for Assisted Reproductive Technology)
- Vitrification
- Embryo Biopsy workshops in our Center of Excellence, Livingston New Jersey
- Embryo Biopsy
- Embryo Mosaicism
- Embryo Transfer
- Genetic Counselling
- EngagedMD

 My Clinic is in the United States
My Clinic is in the United States My Clinic is in Canada
My Clinic is in Canada
